 |
 |
|
 |
|
 |
|  |
|  |
|
 |
|
 |
|  |
|  |
|
 |
A recent post by user 'yesbird' described using a .df3 media-density file made
from a medical CT scan, and rendered in POV-ray...
https://news.povray.org/povray.advanced-users/thread/%3C66f2f986%40news.povray.org%3E/?mtop=444143
I was inspired by that to try a different method, just to see if it would work:
skipping the CT-scan-to-.df3 conversion step and instead using the 'stack' of
multiple CT-scan images themselves as the media density, for the full 3-D
volume.
[I had to use one of the 3.8 betas for this test, because of a limitation in
'official' 3.7.0 of a maximum 256 entries in a pigment_map.]
For the test, I used a CT scan of my own, acquired from a recent visit to my
dentist (for a 'root canal' procedure-- which was painless, I am happy to say!)
In one of the file folders on the CD, I found the original numbered grayscale
image 'slices' that make up the scan-- 358 of them, at 523 X 523 pixel
resolution. Unfortunately, those 16-bit images were in a .dcm file format that
POV-ray cannot directly read as image_maps, so I batch-converted them to .png
files in Irfanview. (That app has a built-in dcm-to-png conversion filter,
although it is not turned on by default.) The one drawback is that the resulting
files were 'down-converted' to 8-bit grayscale...but that was adequate for my
test. The CT scan itself was of rather low-quality anyway.
Image_maps (or 'pigments' in general) cannot be used directly as media density--
but a *function* of the image_maps can. The trick was to get all of the 358
images loaded into a single pigment_map, for conversion to a function; I used an
automated 'string' set-up to do that, in a #for loop. (A typical pigment_map
uses index values between 0.0 and 1.0, which corresponds to 1 unit in z-space by
default. The 358 images fill that volume.)
pigment{
gradient z
pigment_map{
#for(i,1,358)
[i/358
image_map{
png concat("3DSlice",str(i,0,0),".png") gamma 3 interpolate 2 once
}
]
// a higher than normal gamma has the effect of increasing the
// image contrast
#end
.....
It produces the same result as laboriously specifying the individual images
themselves:
[1/358 image_map{png "3DSlice1.png" gamma 3 interpolate 2 once]
[2/358 image_map{png "3DSlice2.png" gamma 3 interpolate 2 once]
.....
[357/358 image_map{png "3DSlice357.png" gamma 3 interpolate 2 once]
[358/358 image_map{png "3DSlice358.png" gamma 3 interpolate 2 once]
(Just a reminder: The images need to be in a file folder location that POV-ray
can find; an additional Library_Path can be added to your master povray.ini
file if needed.)
To be honest, I was not sure *what* the end result of this media scheme would
look like, because of the image_maps being 'stacked together' in z, AND with
small gaps between their z-spacing. (They are just 2-D images.) But in effect,
they produce a kind of 'fake' .df3 appearance-- with a .df3's 'voxel depth'
being replaced by each image's media inbetween the image slices. Apparently,
each image's z-depth media extends only until the next image_map is
encountered-- instead of creating one ugly media blur in the enclosing box.
In the media code, a color_map can be used to give different colors to the
different 'densities'-- meaning, the varying grayscale values of the image_maps.
And the map's colors and index values can be played with to get interesting
effects, like supressing certain densities while accentuating others.
I will also post an animation of the rotating media box, at p.b.f. animations
in the newsgroups.
----
The one unknown value was the 'spacing' of the individual images in the CT
scan-- the CT machine's spatial resolution as the x-ray slices progressed
through my teeth! I just assumed that a 1-unit cube (a box) would suffice as the
full 'depth', more or less. (I actually used <523,523,358>/523 for the scaling)
----
My code:
#version 3.8;
global_settings{assumed_gamma 1.0}
// #default{finish{ambient .05 emission 0 diffuse .9}}
camera {
perspective
location <0, 0, -1.1>
look_at <0, 0, 0>
right x*image_width/image_height
angle 67
}
#declare PIG_SLICES =
function{
pigment{
gradient z
pigment_map{
#for(i,1,358) // 358 images
[i/358
image_map{png concat("3DSlice",str(i,0,0),".png") gamma 3 interpolate 2 once}
]
#end
} // end of pigment_map
} // end of pigment
} // end of function
box{0,.999 translate .0005
hollow
pigment{rgbt 1}
interior{
media{
emission 4
//absorption...
//scattering...
method 3 intervals 1 samples 60 aa_level 4
density{
function{PIG_SLICES(x,y,z).gray} // or .red or .green or .blue
color_map{
[0 transmit 1]
[.01 transmit 1]
[.1 rgb <0,0,1>]
[.2 rgb <0,1,0>]
[.5 rgb <0,1,0>]
[.5 rgb <1,0,0>]
[1 rgb <1,0,0>]
}
} // end of density
} // end of media
} // end of interior
translate -.5
scale <523,523,358>/523
//rotate...
}
Post a reply to this message
Attachments:
Download 'ct_scan_to_media_density_1_kw_10_2024.jpg' (226 KB)
Preview of image 'ct_scan_to_media_density_1_kw_10_2024.jpg'

|
 |
|  |
|  |
|
 |
From: yesbird
Subject: Re: multiple image_maps as media density-- 'fake' .df3
Date: 27 Oct 2024 05:41:02
Message: <671e0aae@news.povray.org>
|
|
 |
|  |
|  |
|
 |
On 27/10/2024 00:36, Kenneth wrote:
> A recent post by user 'yesbird' described using a .df3 media-density file made
> from a medical CT scan, and rendered in POV-ray...
Brilliant !
This is a very interesting and unusual experiment with surprisingly
excellent results - I even didn't know about such techniques. Looks like
now we can skip one step in preparing media data - it's very promising.
I did it with following commands before:
$ convert image-000001.dcm imgs.png
$ df3util create 512,1024,128 1 test
$ for i in $(seq 0 127) ; do df3util import test z:$i
$(printf "imgs-%d.png" $i) ; done
'df3util' written by jr works fine, but ImageMagic's 'convert' on
some platforms gives error processing jpeg, so I switched to:
$ dcmj2pnm image-000001.dcm --write-png > image-000001.png from dcmtk
package (sudo apt-get install dcmtk).
Maybe this utils will preserve 16-but color depth, I didn't check it.
Now there are two questions of interest for me:
1. Quality comparison of images, produced by both methods. I see that
color gradient on your images is not very smooth, compared to
traditional method, but maybe this is colormap dependent.
2. How it will work for isosurface approach (see attached
image): https://povlab.online/?scene=dicom_iso.pov
I am continuing to think about a volume rendering service with Three.js
based GUI with POV on the server side and your method can simplify
implementation if we will not see quality degradation. In general DICOM
is a great source of data for volumes, thanks for displaying your
teeths, hope you will not need this operation in future.
--
YB
Post a reply to this message
Attachments:
Download 'dicom_iso.png' (97 KB)
Preview of image 'dicom_iso.png'

|
 |
|  |
|  |
|
 |
|
 |
|  |
|  |
|
 |
yesbird <sya### [at] gmail com> wrote:
>
> color gradient on your images is not very smooth, compared to
> traditional method, but maybe this is colormap dependent.
Your media images from your earlier post definitely look much nicer than mine.
The main problem I am having is that the original DICOM images from my own CT
scan are *extremely* noisy and of low quality; it is even difficult to see what
is 'organic matter' and what is just noise. And the exposure level from image to
image seems to change at random, which I suspect is a result of the CT's x-rays
being 'auto-compensated' for changes in bone density (or whatever!)
The color_map I used (and the altered gamma setting for the images) was the
result of a lot of experimentation, to see if I could suppress some of the noisy
'garbage' and still retain some of the important details and structures. Small
tweaks to the color_map produce large changes in the visual appearance!
I have attached a sample of my original DICOM images (as converted to
8-bit .png) just to show what the problems are. I think that the images you are
using are of *much* higher quality.
In my opinion, my dentist needs to purchase an improved CT scanner! ;-)
>
> 2. How it will work for isosurface approach?
I am working on that at the moment (thanks for the suggestion!) I will post the
result when finished.
>
> hope you will not need this operation in future.
I hope so too-- mainly because the procedure is EXPENSIVE! $$$ com> wrote:
>
> color gradient on your images is not very smooth, compared to
> traditional method, but maybe this is colormap dependent.
Your media images from your earlier post definitely look much nicer than mine.
The main problem I am having is that the original DICOM images from my own CT
scan are *extremely* noisy and of low quality; it is even difficult to see what
is 'organic matter' and what is just noise. And the exposure level from image to
image seems to change at random, which I suspect is a result of the CT's x-rays
being 'auto-compensated' for changes in bone density (or whatever!)
The color_map I used (and the altered gamma setting for the images) was the
result of a lot of experimentation, to see if I could suppress some of the noisy
'garbage' and still retain some of the important details and structures. Small
tweaks to the color_map produce large changes in the visual appearance!
I have attached a sample of my original DICOM images (as converted to
8-bit .png) just to show what the problems are. I think that the images you are
using are of *much* higher quality.
In my opinion, my dentist needs to purchase an improved CT scanner! ;-)
>
> 2. How it will work for isosurface approach?
I am working on that at the moment (thanks for the suggestion!) I will post the
result when finished.
>
> hope you will not need this operation in future.
I hope so too-- mainly because the procedure is EXPENSIVE! $$$
Post a reply to this message
Attachments:
Download 'four_sample_dicom_images_as_8_bit_png.jpg' (109 KB)
Preview of image 'four_sample_dicom_images_as_8_bit_png.jpg'
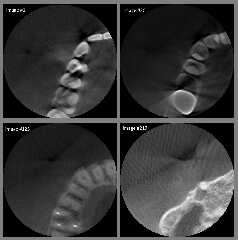
|
 |
|  |
|  |
|
 |
From: yesbird
Subject: Re: multiple image_maps as media density-- 'fake' .df3
Date: 27 Oct 2024 14:39:39
Message: <671e88eb$1@news.povray.org>
|
|
 |
|  |
|  |
|
 |
On 27/10/2024 19:00, Kenneth wrote:
> The main problem I am having is that the original DICOM images from my own CT
> scan are *extremely* noisy and of low quality; it is even difficult to see what
> is 'organic matter' and what is just noise.
Yes, looking at attached images I wonder how dentist can fetch some
useful information from them.
> The color_map I used (and the altered gamma setting for the images) was the
> result of a lot of experimentation, to see if I could suppress some of the noisy
> 'garbage' and still retain some of the important details and structures. Small
> tweaks to the color_map produce large changes in the visual appearance!
Yes, turning colormaps can be painful, but I can suggest following:
Don't know what environment are you using, but this extension to VS-Code
has a very useful colormaps editor:
https://github.com/VirtualWhirlwind/vscode-povray2
Also a collection of prepared colormaps in my repository can be used as
a good starting point:
https://github.com/syanenko/pov-colormaps
> In my opinion, my dentist needs to purchase an improved CT scanner! ;-)
Definitely, and share the images with us :)).
> I am working on that at the moment (thanks for the suggestion!) I will post the
> result when finished.
Very interesting - looking forward to see them.
--
YB
Post a reply to this message
|
 |
|  |
|  |
|
 |
|
 |
|  |
|  |
|
 |
"Kenneth" <kdw### [at] gmail com> wrote:
> yesbird <sya### [at] gmail com> wrote:
> yesbird <sya### [at] gmail com> wrote:
> >
> > 2. How it will work for isosurface approach?
>
> I am working on that at the moment (thanks for the suggestion!) I will post the
> result when finished.
>
Here is the isosurface example, using the same series of images. It worked,
although I had difficulty with the noise in the original images.
The PIG_SLICES function code is the same as before, but instead of using it for
media, I used...
isosurface {
function {1.19 - PIG_SLICES(x,y,z).red}
contained_by {box {.001, .999}}
threshold .65
accuracy .0001
max_gradient 390
texture{...}
}
I chose the various values-- especially the 1.19-- to try and eliminate as much
of the noise as possible...without wiping out *too* much of the detail. Then I
rotated the isosurface into a more visually interesting orientation.
Surprisingly, it does not show a 'stair-step' appearance from the individual
image_maps; they are all nicely blended together.
Thanks yesbird for the inspiration! While working on this, I discovered
something important (and off-topic) that has given me big problems in the past,
concerning the fact that this isosurface actually responds to *lighting*. I was
not expecting that. The reason why is worth a separate post, if I can get around
to it. ;-) com> wrote:
> >
> > 2. How it will work for isosurface approach?
>
> I am working on that at the moment (thanks for the suggestion!) I will post the
> result when finished.
>
Here is the isosurface example, using the same series of images. It worked,
although I had difficulty with the noise in the original images.
The PIG_SLICES function code is the same as before, but instead of using it for
media, I used...
isosurface {
function {1.19 - PIG_SLICES(x,y,z).red}
contained_by {box {.001, .999}}
threshold .65
accuracy .0001
max_gradient 390
texture{...}
}
I chose the various values-- especially the 1.19-- to try and eliminate as much
of the noise as possible...without wiping out *too* much of the detail. Then I
rotated the isosurface into a more visually interesting orientation.
Surprisingly, it does not show a 'stair-step' appearance from the individual
image_maps; they are all nicely blended together.
Thanks yesbird for the inspiration! While working on this, I discovered
something important (and off-topic) that has given me big problems in the past,
concerning the fact that this isosurface actually responds to *lighting*. I was
not expecting that. The reason why is worth a separate post, if I can get around
to it. ;-)
Post a reply to this message
Attachments:
Download 'image_stack_as_isosurface_1_kw.jpg' (102 KB)
Preview of image 'image_stack_as_isosurface_1_kw.jpg'

|
 |
|  |
|  |
|
 |
From: yesbird
Subject: Re: multiple image_maps as media density-- 'fake' .df3
Date: 2 Nov 2024 05:05:00
Message: <6725eb3c@news.povray.org>
|
|
 |
|  |
|  |
|
 |
On 01/11/2024 16:05, Kenneth wrote:
> Here is the isosurface example, using the same series of images. It worked,
> although I had difficulty with the noise in the original images.
Yes, I know - sometimes even in real life it is very difficult to
separate useful info from noise :).
>
> Surprisingly, it does not show a 'stair-step' appearance from the individual
> image_maps; they are all nicely blended together.
Really interesting and unexpected - in general even with such low
resolution image looks smoothly. The noisy sources sometimes produces
very impressive effects - in this particular case it reminds me old VHS
fantastic movies, something like "Spiders from Mars" or "War of the
Worlds" (see attached image with post-production effect :)).
>
> Thanks yesbird for the inspiration! While working on this, I discovered
> something important (and off-topic) that has given me big problems in the past,
> concerning the fact that this isosurface actually responds to *lighting*. I was
> not expecting that. The reason why is worth a separate post, if I can get around
> to it. ;-)
Looking forward to look at your future investigations.
--
YB
Post a reply to this message
Attachments:
Download 'image_stack_as_isosurface_1_kw_pp.tif.dat' (4727 KB)
|
 |
|  |
|  |
|
 |
From: Paolo Gibellini
Subject: Re: multiple image_maps as media density-- 'fake' .df3
Date: 8 Dec 2024 17:07:01
Message: <67561885$1@news.povray.org>
|
|
 |
|  |
|  |
|
 |
Il 27/10/2024 17:00, Kenneth ha scritto:
> yesbird <sya### [at] gmail com> wrote:
>>
[...]
Great results!
> In my opinion, my dentist needs to purchase an improved CT scanner! ;-)
Usually dentist use low-dose CT scanner like the Cone Beam or similar,
and the spatial resolution is lower than the CT scans used in hospitals.
Paolo com> wrote:
>>
[...]
Great results!
> In my opinion, my dentist needs to purchase an improved CT scanner! ;-)
Usually dentist use low-dose CT scanner like the Cone Beam or similar,
and the spatial resolution is lower than the CT scans used in hospitals.
Paolo
Post a reply to this message
|
 |
|  |
|  |
|
 |
|
 |
|  |
|  |
|
 |
Paolo Gibellini <p.g### [at] gmail com> wrote:
>
> Great results!
>
Thanks, Paolo. I only wish that the DICOM image quality had been better.
>
> > In my opinion, my dentist needs to purchase an improved CT scanner! ;-)
>
>
> Usually dentist use low-dose CT scanner like the Cone Beam or similar,
> and the spatial resolution is lower than the CT scans used in hospitals.
>
Ah, that explains it. Thanks, I have learned something new!
(I recently had several x-rays made of my spine-- although not a CT-scan-- and
got copies of them from the doctor. They are very high-resolution!) com> wrote:
>
> Great results!
>
Thanks, Paolo. I only wish that the DICOM image quality had been better.
>
> > In my opinion, my dentist needs to purchase an improved CT scanner! ;-)
>
>
> Usually dentist use low-dose CT scanner like the Cone Beam or similar,
> and the spatial resolution is lower than the CT scans used in hospitals.
>
Ah, that explains it. Thanks, I have learned something new!
(I recently had several x-rays made of my spine-- although not a CT-scan-- and
got copies of them from the doctor. They are very high-resolution!)
Post a reply to this message
|
 |
|  |
|  |
|
 |
From: Paolo Gibellini
Subject: Re: multiple image_maps as media density-- 'fake' .df3
Date: 26 Dec 2024 14:01:37
Message: <676da811@news.povray.org>
|
|
 |
|  |
|  |
|
 |
12/12/2024 04:43, Kenneth wrote:
> (I recently had several x-rays made of my spine-- although not a CT-scan-- and
> got copies of them from the doctor. They are very high-resolution!)
>
(Yes, the technology used in traditional X-Ray exams, like in Direct
Radiography, allows a better resolution due to a lower pixel size. New
Photon-counting CT will probably allow better volumetric resolution, it
is a fascinating world!)
Post a reply to this message
|
 |
|  |
|  |
|
 |
|
 |
|  |
|
 |




![]()